Merge branch 'release2.3' into 'develop'
Release2.3 See merge request !606
No related branches found
No related tags found
Showing
- .github/workflows/main.yml 6 additions, 6 deletions.github/workflows/main.yml
- docs/Makefile 0 additions, 3 deletionsdocs/Makefile
- docs/cookbooks/MultivariateFlagging.rst 5 additions, 5 deletionsdocs/cookbooks/MultivariateFlagging.rst
- docs/requirements.txt 1 addition, 2 deletionsdocs/requirements.txt
- docs/resources/data/hydro_config.csv 3 additions, 3 deletionsdocs/resources/data/hydro_config.csv
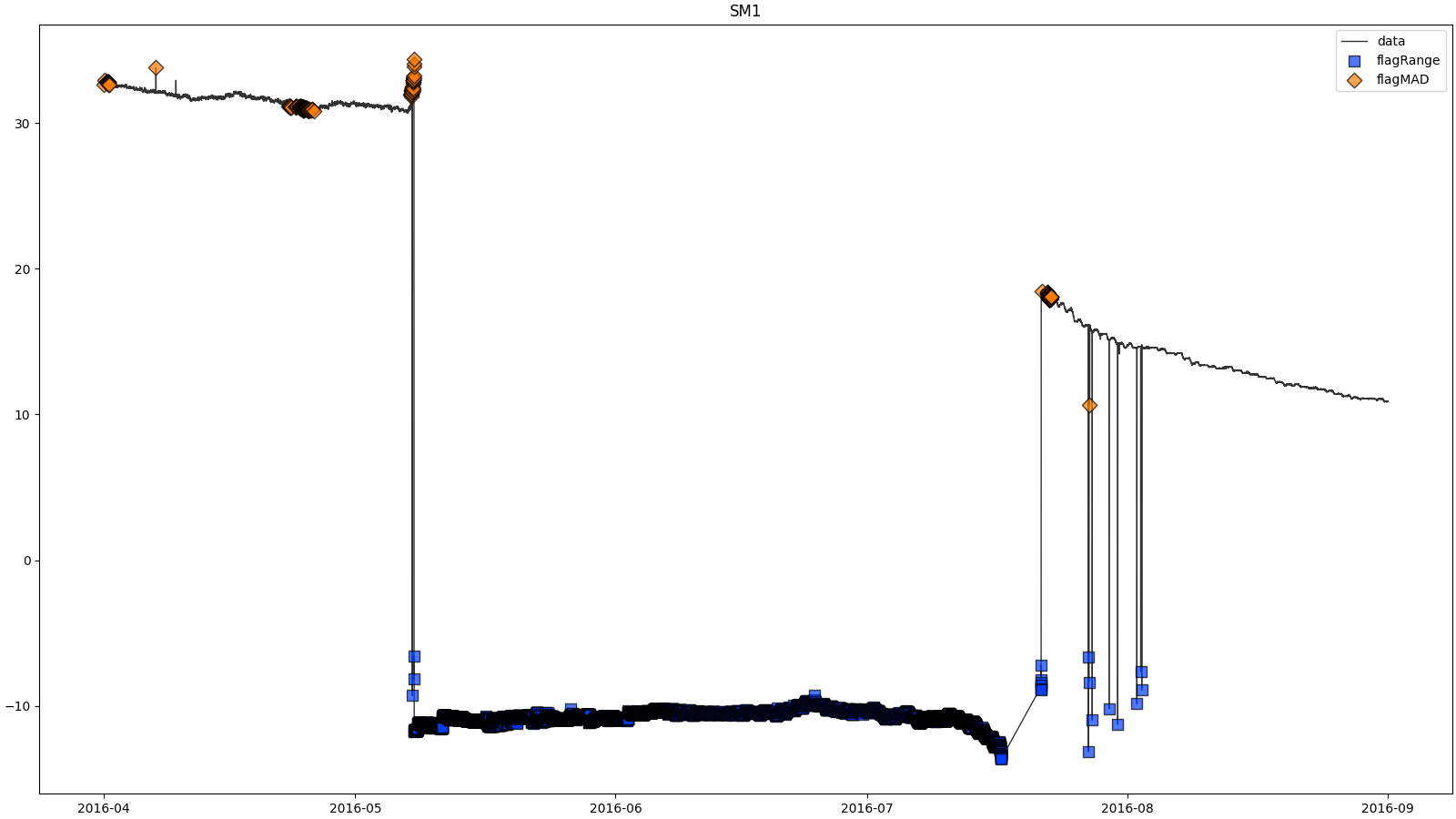
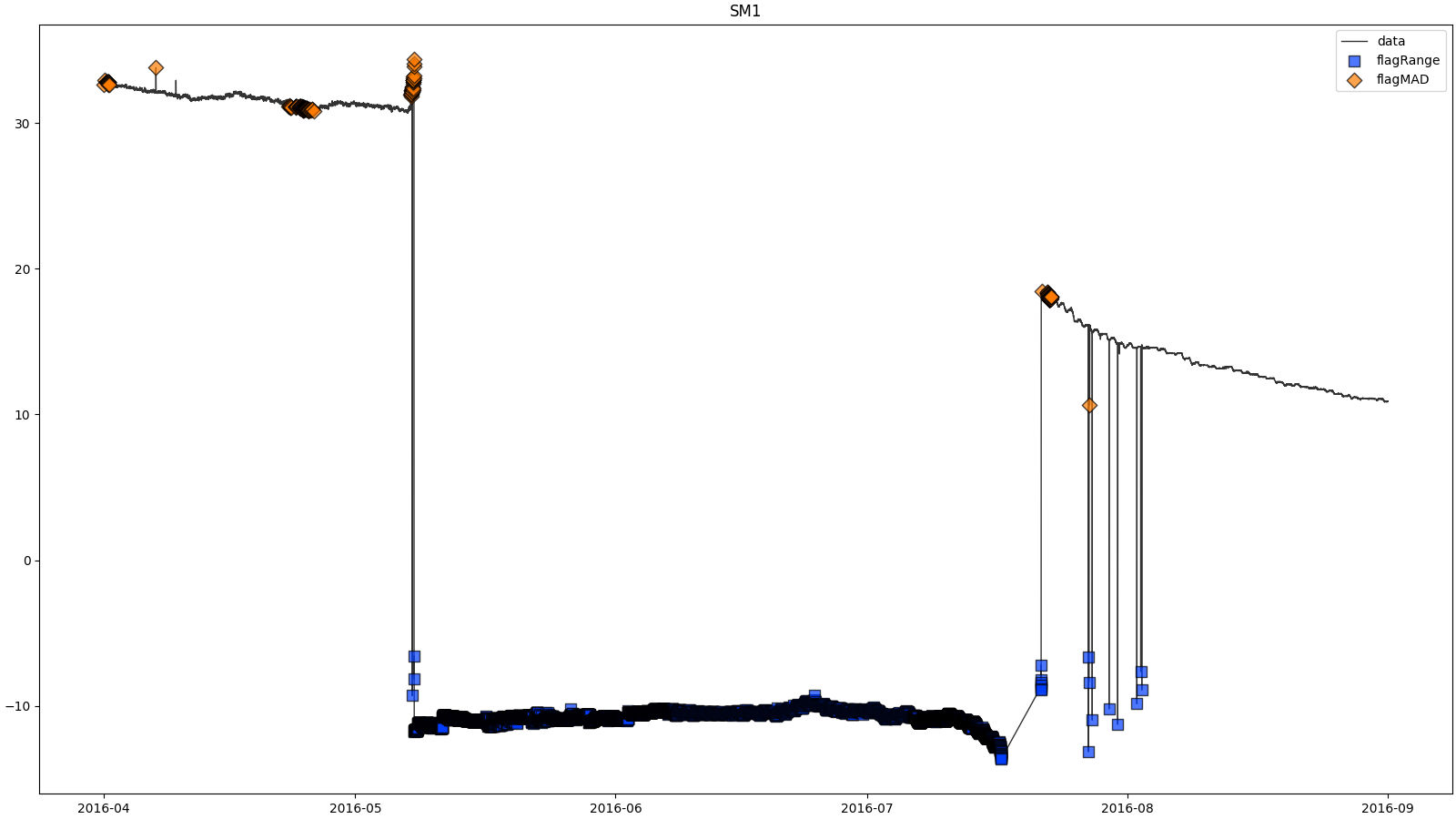
- docs/resources/temp/SM1processingResults.png 0 additions, 0 deletionsdocs/resources/temp/SM1processingResults.png
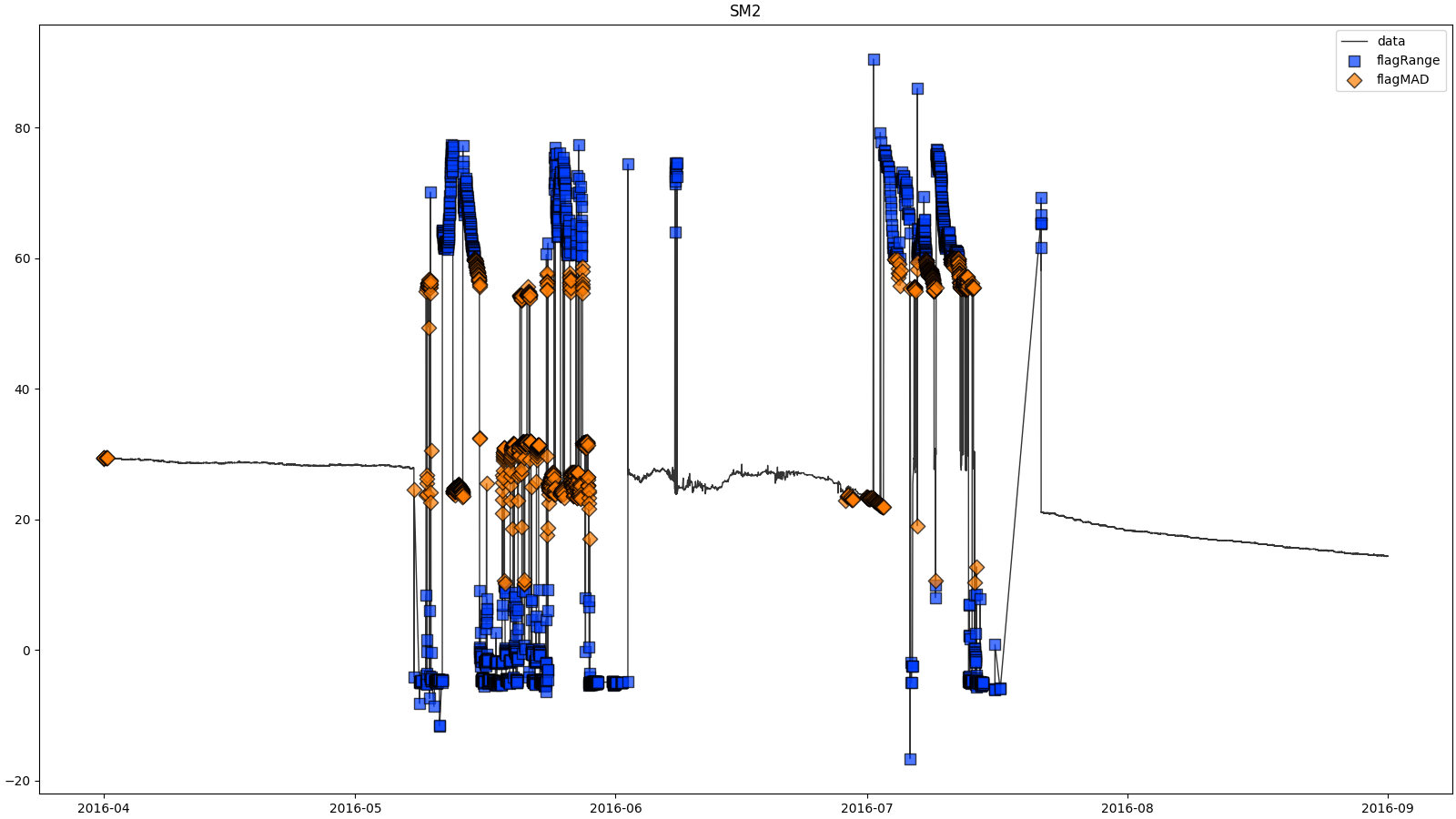
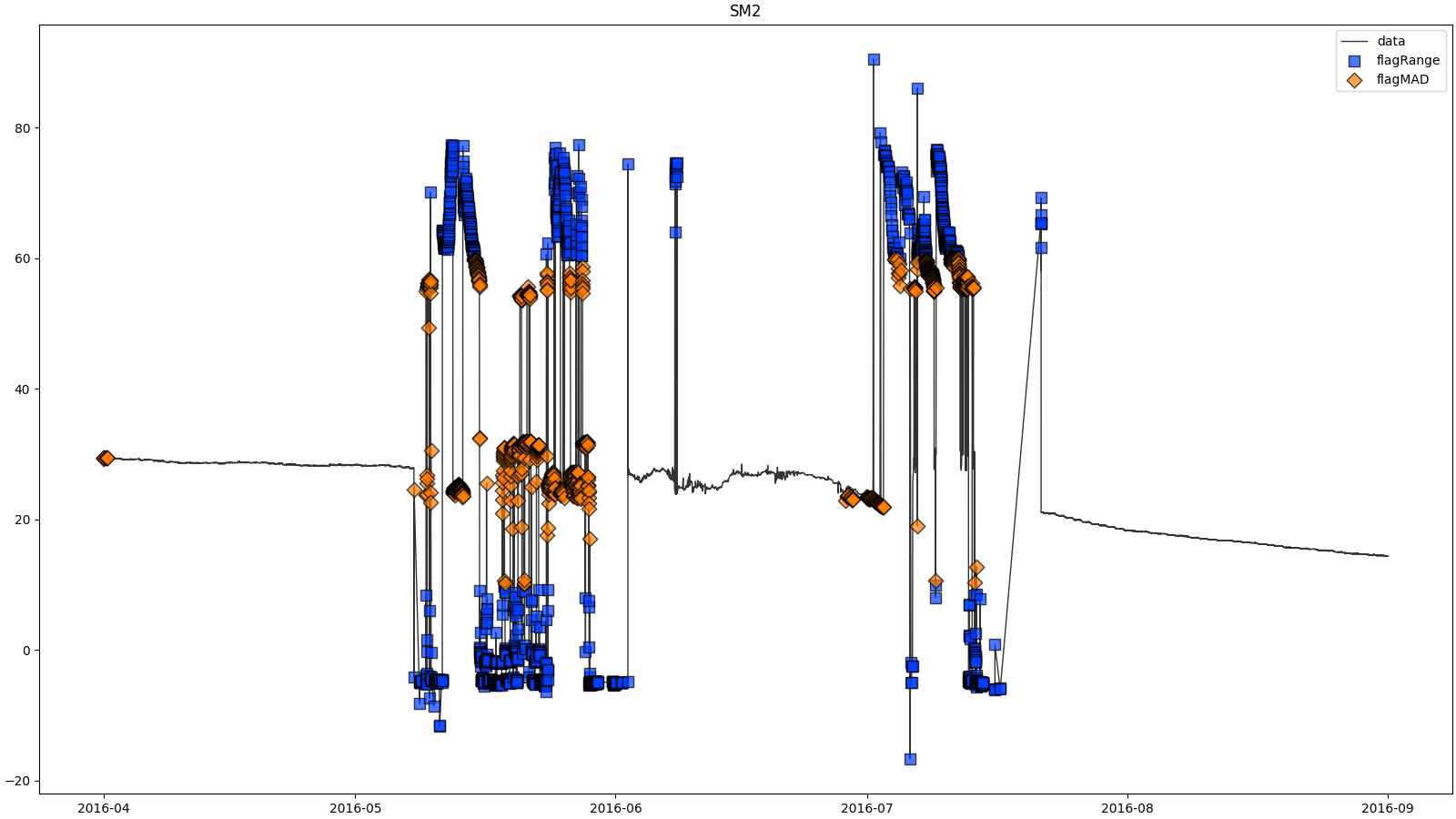
- docs/resources/temp/SM2processingResults.png 0 additions, 0 deletionsdocs/resources/temp/SM2processingResults.png
| W: | H:
| W: | H:


| W: | H:
| W: | H: