No related branches found
No related tags found
Showing
- .Rbuildignore 1 addition, 0 deletions.Rbuildignore
- .gitattributes 1 addition, 0 deletions.gitattributes
- .gitignore 0 additions, 1 deletion.gitignore
- DESCRIPTION 3 additions, 3 deletionsDESCRIPTION
- README.md 62 additions, 25 deletionsREADME.md
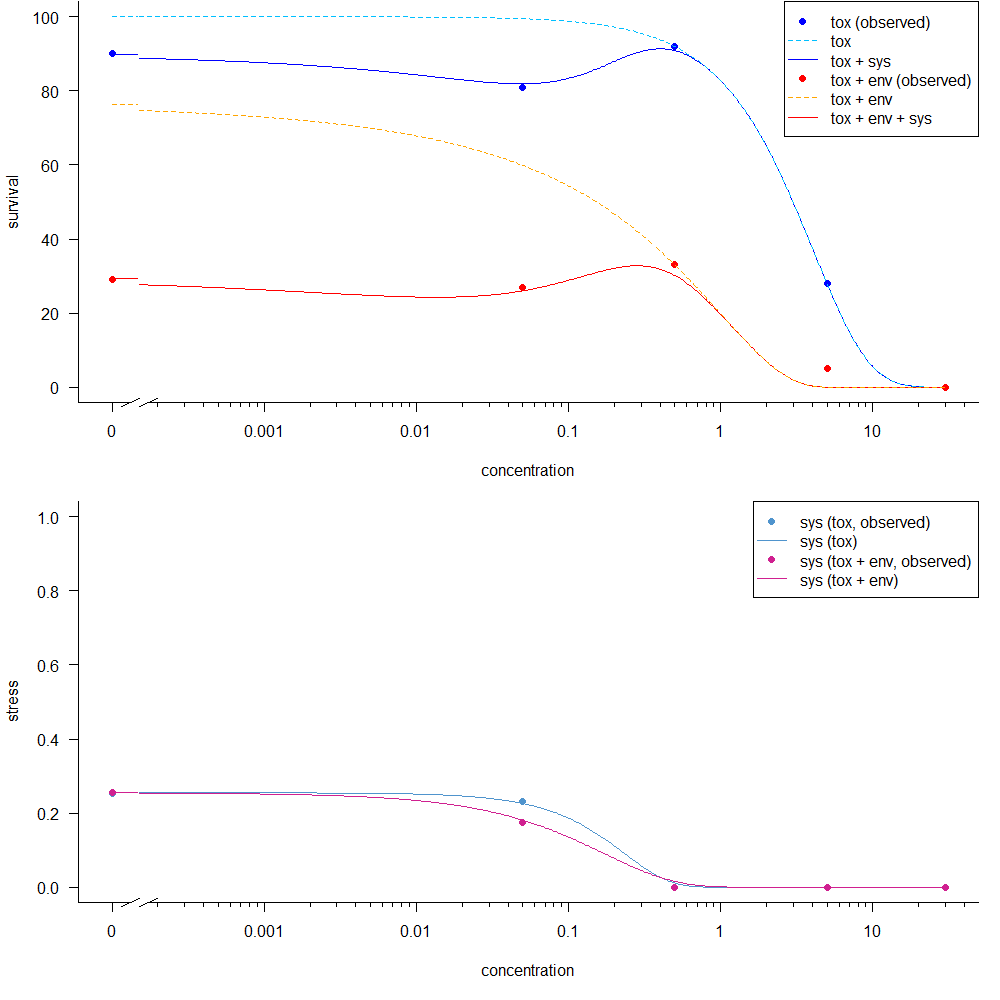
- images/example.png 3 additions, 0 deletionsimages/example.png
- man/stressaddition-package.Rd 1 addition, 1 deletionman/stressaddition-package.Rd
.gitattributes
0 → 100644
130 B