Commits on Source (31)
-
David Schäfer authored09d7be17
-
David Schäfer authored
Update and fix plot docstring See merge request !696
84074146 -
Peter Lünenschloß authored9f61bcee
-
Peter Lünenschloß authored
Merge z scoring methods See merge request !691
249ccbcd -
Peter Lünenschloß authoredc9d73c2d
-
Peter Lünenschloß authored
Revert "Merge branch 'mergeZScoringMethods' into 'develop'" See merge request !699
b0d9e815 -
David Schäfer authored18a19948
-
David Schäfer authored2a92c466
-
David Schäfer authored
added deprecation warnings to the docstrings See merge request !701
f4507dac -
Bert Palm authored3ff66907
-
eb3f9d73
-
David Schäfer authored
Bump hypothesis from 6.72.2 to 6.75.1 See merge request !689
cd1ac6ba -
Bert Palm authoredd698e8d2
-
Bert Palm authored04e7b524
-
Peter Lünenschloß authoredd873be8a
-
Peter Lünenschloß authored
Drift flagging cookbook See merge request !675
c6bdd8b3 -
David Schäfer authorede566c729
-
David Schäfer authored
release2.4.1 See merge request !707
c474f18b -
David Schäfer authored2d7d1c1f
-
David Schäfer authored
Release2.4.1 See merge request !708
9fce4593 -
Peter Lünenschloß authored21f74914
-
Peter Lünenschloß authored
Merge z scoring methods See merge request !700
85cc0535 -
Peter Lünenschloß authored5ec45a5b
-
Peter Lünenschloß authored
Numba riddance See merge request !703
1249d3b1 -
Bert Palm authoredb1a4dc1e
-
Peter Lünenschloß authored1349ad0d
-
Peter Lünenschloß authored
removed debug artifact See merge request !711
53c7e616
Showing
- .gitlab-ci.yml 28 additions, 1 deletion.gitlab-ci.yml
- CHANGELOG.md 19 additions, 3 deletionsCHANGELOG.md
- README.md 2 additions, 2 deletionsREADME.md
- docs/cookbooks/CookBooksPanels.rst 11 additions, 0 deletionsdocs/cookbooks/CookBooksPanels.rst
- docs/cookbooks/DriftDetection.rst 233 additions, 0 deletionsdocs/cookbooks/DriftDetection.rst
- docs/cookbooks/ResidualOutlierDetection.rst 2 additions, 2 deletionsdocs/cookbooks/ResidualOutlierDetection.rst
- docs/misc/title.rst 1 addition, 0 deletionsdocs/misc/title.rst
- docs/resources/data/config.csv 1 addition, 1 deletiondocs/resources/data/config.csv
- docs/resources/data/config_ci.csv 1 addition, 1 deletiondocs/resources/data/config_ci.csv
- docs/resources/data/myconfig.csv 1 addition, 1 deletiondocs/resources/data/myconfig.csv
- docs/resources/data/myconfig2.csv 1 addition, 1 deletiondocs/resources/data/myconfig2.csv
- docs/resources/data/myconfig4.csv 2 additions, 2 deletionsdocs/resources/data/myconfig4.csv
- docs/resources/data/tempSensorGroup.csv 52560 additions, 0 deletionsdocs/resources/data/tempSensorGroup.csv
- docs/resources/data/tempSensorGroup.csv.license 3 additions, 0 deletionsdocs/resources/data/tempSensorGroup.csv.license
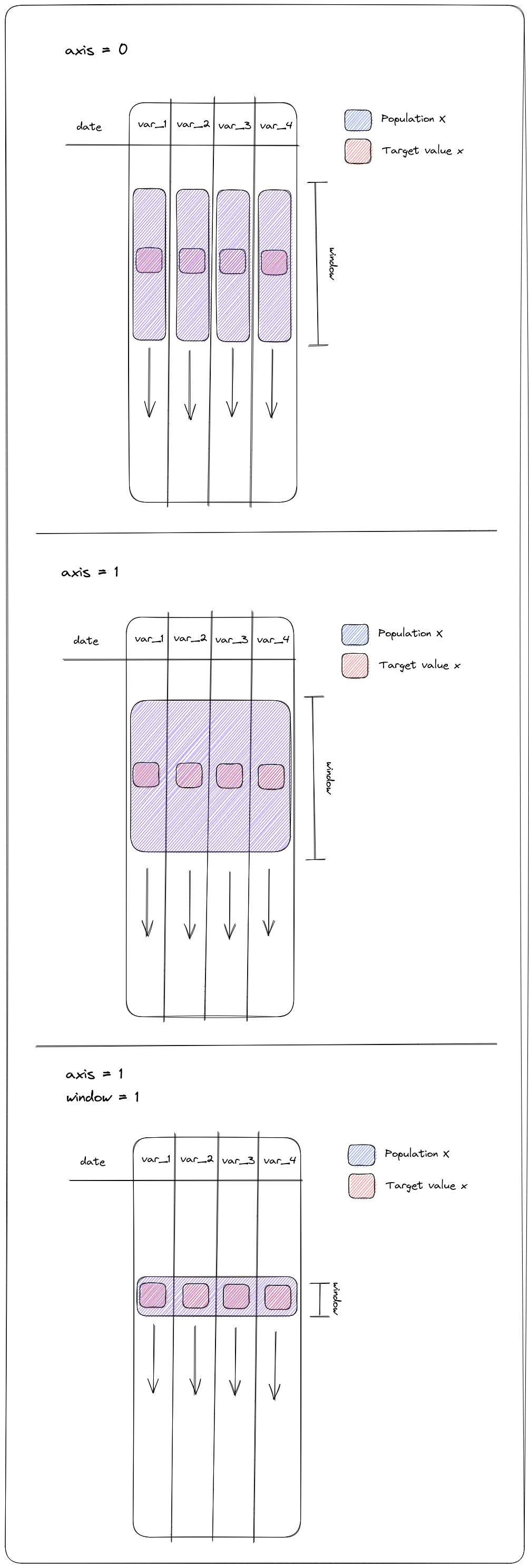
- docs/resources/images/ZscorePopulation.png 0 additions, 0 deletionsdocs/resources/images/ZscorePopulation.png
- docs/resources/images/ZscorePopulation.png.license 3 additions, 0 deletionsdocs/resources/images/ZscorePopulation.png.license
- requirements.txt 1 addition, 2 deletionsrequirements.txt
- saqc/core/flags.py 0 additions, 1 deletionsaqc/core/flags.py
- saqc/core/frame.py 0 additions, 30 deletionssaqc/core/frame.py
- saqc/funcs/changepoints.py 8 additions, 44 deletionssaqc/funcs/changepoints.py
docs/cookbooks/DriftDetection.rst
0 → 100644
docs/resources/data/tempSensorGroup.csv
0 → 100644
This diff is collapsed.
docs/resources/images/ZscorePopulation.png
0 → 100644
793 KiB
| ... | ... | @@ -6,7 +6,6 @@ Click==8.1.3 |
| docstring_parser==0.15 | ||
| dtw==1.4.0 | ||
| matplotlib==3.7.1 | ||
| numba==0.57.0 | ||
| numpy==1.24.3 | ||
| outlier-utils==0.0.3 | ||
| pyarrow==11.0.0 | ||
| ... | ... | @@ -14,4 +13,4 @@ pandas==2.0.1 |
| scikit-learn==1.2.2 | ||
| scipy==1.10.1 | ||
| typing_extensions==4.5.0 | ||
| fancy-collections==0.1.3 | ||
| fancy-collections==0.2.1 | ||
| \ No newline at end of file |